Introduction
Artificial intelligence (AI) has transformed many fields, from image recognition to drug discovery, yet its integration into biomedical research remains slower than anticipated. While discussions often center around computational power, algorithms, and cloud infrastructure, the real challenge is far more fundamental: data quality and annotation.
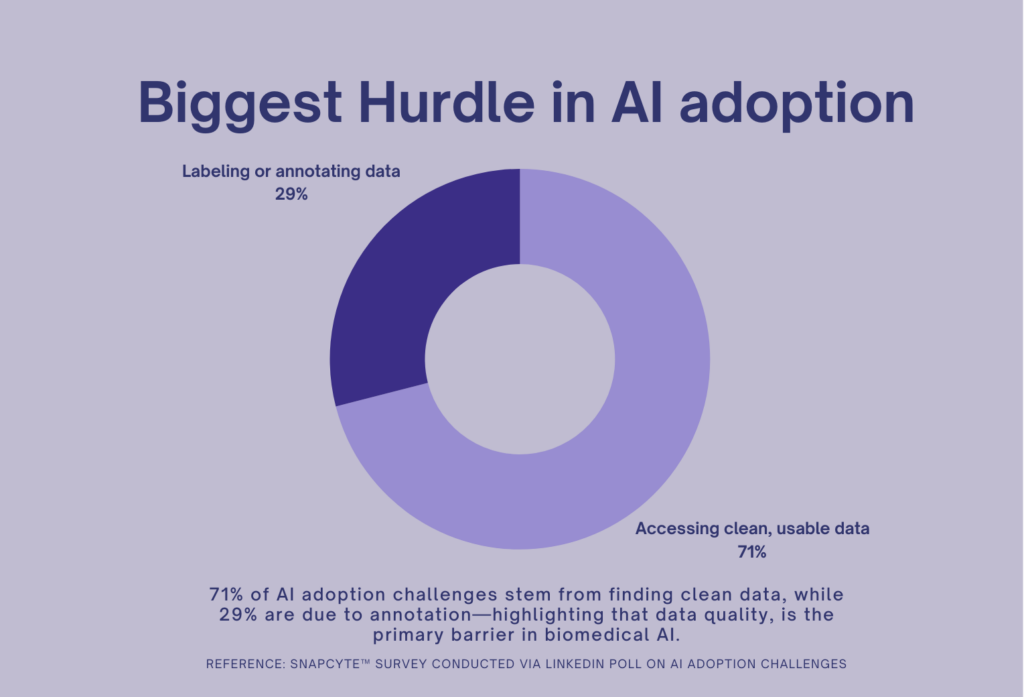
A recent poll we conducted reinforced this reality—71% of respondents identified finding clean data as the biggest hurdle, while 29% pointed to data annotation. This highlights a crucial issue: AI adoption in biomedical research is bottlenecked not by technology, but by the data itself.
Why Biomedical AI Struggles with Data
AI models thrive on structured, high-quality data. However, biomedical research presents unique challenges that make AI integration particularly difficult.
1. Biomedical Data Is Messy and Heterogeneous
Unlike standardized datasets in finance or natural language processing, biomedical data comes in many forms: microscopy images, genomic sequences, patient records, and multi-modal datasets from different instruments. There is no single standard governing how these datasets should be stored, labeled, or structured. According to Han (2025), one of the biggest obstacles to reproducible AI in biomedical research is the inherent conflict between standardization and researchers’ individualized data handling practices. As a result, simply acquiring a dataset is not enough—it needs extensive preprocessing, filtering, and standardization before AI can make sense of it.
2. Scientists Are Not Data Scientists
Many biomedical researchers are highly skilled in their respective domains but lack formal training in data science or machine learning. A common problem in academic and biotech labs is suboptimal data storage and organization—files are often stored in fragmented formats, with inconsistent metadata and minimal documentation (Wilkinson et al., 2016).
This gap leads to poor reproducibility and makes datasets difficult to integrate into AI workflows. As AI adoption grows, the need for better data curation strategies and training for scientists in data handling is becoming increasingly evident.
3. Annotation is Expensive, Time-Consuming, and Requires Expertise
Data annotation is a well-documented bottleneck in AI training. For biomedical datasets, annotations often require domain-specific knowledge—a task that cannot be outsourced to non-experts without compromising accuracy. In medical imaging, for instance, expert-labeled data is essential for training diagnostic models, but the annotation process is slow and resource-intensive (Litjens et al., 2017).
The challenge extends beyond simple labeling. Variability in annotations due to subjective interpretation introduces bias into AI models, making it difficult to achieve reliable outcomes across different datasets.
The Path Forward: Addressing Data Challenges in AI-Driven Research
The biomedical AI community is actively seeking solutions to these problems, including:
•Developing Standardized Data Repositories: Initiatives like the FAIR Principles (Findability, Accessibility, Interoperability, and Reusability) aim to create structured, well-documented datasets (Wilkinson et al., 2016).
•Automated Preprocessing and Cleaning Pipelines: AI-driven tools are being developed to clean and structure biomedical datasets before they are used for training (Rajkomar et al., 2019).
•Better Training for Researchers: Increasing awareness and providing accessible training on data management, annotation, and AI best practices will be crucial in bridging the gap between biomedical science and machine learning.
Final Thoughts
AI has immense potential to revolutionize biomedical research, but it cannot do so without addressing the fundamental data bottlenecks. The focus must shift from just developing better models to ensuring high-quality, structured, and expertly annotated data. Without this, even the most powerful AI systems will struggle to generate reliable and reproducible results.
At SnapCyte™, we’ve faced these challenges firsthand. Our AI models were trained on over 5,000 expertly annotated microscopy images, ensuring the highest level of accuracy. We now help research teams organize, clean, and annotate their datasets to overcome these roadblocks. But ultimately, this is an industry-wide challenge that needs collective effort.
If you’re facing challenges with data curation, annotation, or structuring for AI applications, contact us to learn how we can help streamline your workflow.
References
Han, H. (2025). Challenges of reproducible AI in biomedical data science. BMC Medical Genomics, 18(8).
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., … & Mons, B. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data, 3, 160018.
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., … & van der Laak, J. A. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60-88.
Rajkomar, A., Dean, J., & Kohane, I. (2019). Machine learning in medicine. New England Journal of Medicine, 380(14), 1347-1358.